Segrefine3dcyclexplore¶
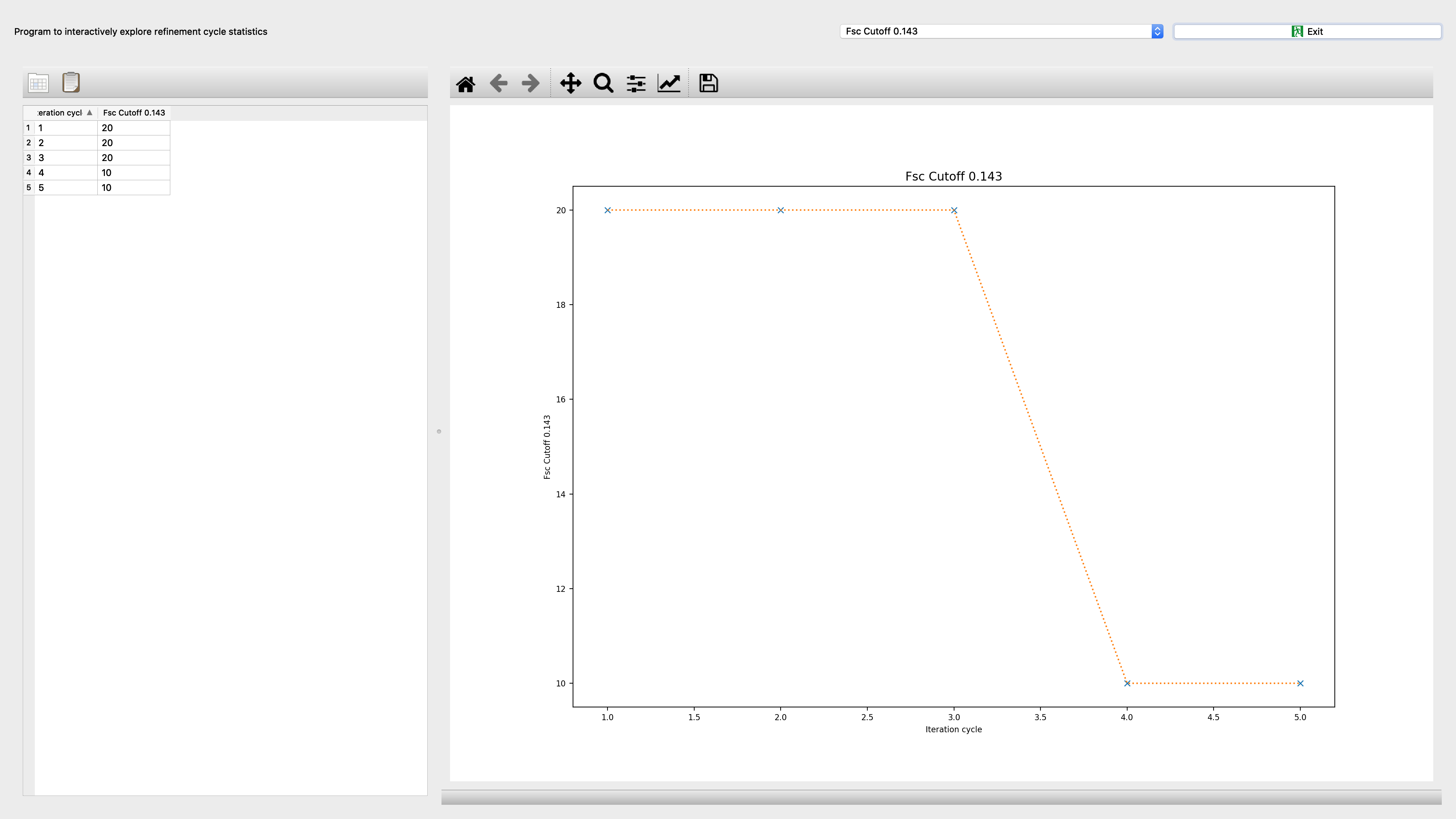
Program to interactively explore refinement cycle statistics
Parameters¶
Parameter |
Example (default) |
Description |
|---|---|---|
refinement.db file |
refinement.db |
Requires refinement.db from segmentrefine3d to extract refinment parameters. |
Batch mode |
False |
Batch mode for plot. Otherwise interactive. |
Sample parameter file¶
You may run the program in the command line by providing the parameters via a text file:
segrefine3dcyclexplore --f parameterfile.txt
Where the format of the parameters is:
refinement.db file = refinement.db
Batch mode = False
Additional parameters (intermediate level)¶
Parameter |
Example (default) |
Description |
|---|---|---|
Diagnostic plot prefix |
diagnostic_plot.pdf |
Output name for diagnostic plots of iterative structure refinement (completion to ‘prefix_XXX.ext’): accepted file formats (pdf, .png, .bmp, .emf, .eps, .gif, .jpeg, .jpg, .ps, .raw, .rgba, .svg, .svgz, .tif, .tiff). |
Criterion |
mean_helical_ccc |
Refinement criteria of refinement to be plotted: ‘fsc_0143’: FSC cutoff 0.143 - Fourier shell correlation at 0.143 cutoff Rosenthal/Henderson criterion in Angstrom.; ‘fsc_05’: FSC cutoff 0.5 - Fourier shell correlation at 0.5 cutoff criterion in Angstrom.; ‘xshift_error’: Shift error perpendicular to helix in Angstrom - derived from RMS deviation of forward difference between successive segments.; ‘inplane_error’: In-plane rotation error perpendicular to helix in degrees - derived from RMS deviation of forward difference between successive segments.; ‘outofplane_error’: Out-of-plane tilt error to helix in degrees - derived from RMS deviation of forward difference between successive segments.; ‘mean_peak’: Mean peak - of projection matching between experimental images and reprojections.; ‘amp_corr_quarter_nyquist’: Average amplitude correlation up to quarter Nyquist cutoff - between experimental and simulated power spectrum.; ‘amp_corr_half_nyquist’: Average amplitude correlation up to half Nyquist cutoff - between experimental and simulated power spectrum.; ‘amp_corr_3quarter_nyquist’: Average amplitude correlation up to three quarter Nyquist cutoff - between experimental and simulated power spectrum.; ‘amp_correlation’: Average amplitude correlation - between experimental and simulated power spectrum.; ‘helical_ccc_error’: Helical CCC error - Mean of standard deviations of CCC values from all asymmetric unit views with their corresponding reprojections.; ‘mean_helical_ccc’: Mean helical CCC - Mean of CCC values from all asymmetric unit views with their corresponding reprojections.; ‘out_of_plane_dev’: Out of plane tilt deviation - Standard deviationof out-of-plane tilt angles ; ‘variance’: Variance - of 3D reconstruction.; ‘asym_unit_count’: Asymmetric unit count - Number of asymmetric units included in 3D reconstruction.; ‘avg_azimuth_sampling’: Azimuthal sampling average - Average of sampling of phi angles around helical axis; ‘dev_azimuth_sampling’: Azimuthal sampling standard deviation - Standard deviation of sampling of phi angles around helical axis; ‘excluded_out_of_plane_tilt_count’: Excluded out-of-plane segment count - based on out-of-plane tilt angle criterion; ‘excluded_inplane_count’: Excluded inplane segment count - based on in-plane rotation angle criterion; ‘total_excluded_count’: Excluded segment count - based on applied refinement criteria; |
Sample parameter file (intermediate level)¶
You may run the program in the command line by providing the parameters via a text file:
segrefine3dcyclexplore --f parameterfile.txt
Where the format of the parameters is:
refinement.db file = refinement.db
Batch mode = False
Diagnostic plot prefix = diagnostic_plot.pdf
Criterion = mean_helical_ccc
Additional parameters (expert level)¶
Parameter |
Example (default) |
Description |
|---|---|---|
Sample parameter file (expert level)¶
You may run the program in the command line by providing the parameters via a text file:
segrefine3dcyclexplore --f parameterfile.txt
Where the format of the parameters is:
refinement.db file = refinement.db
Batch mode = False
Diagnostic plot prefix = diagnostic_plot.pdf
Criterion = mean_helical_ccc
Command line options¶
When invoking segrefine3dcyclexplore, you may specify any of these options:
usage: segrefine3dcyclexplore [-h] [--g] [--p] [--f FILENAME] [--c] [--l LOGFILENAME] [--d DIRECTORY_NAME] [--version] [--batch_mode]
[input_output [input_output ...]]
Program to interactively explore refinement cycle statistics
positional arguments:
input_output Input and output files
optional arguments:
-h, --help show this help message and exit
--g, --GUI GUI option: read input parameters from GUI
--p, --promptuser Prompt user option: read input parameters from prompt
--f FILENAME, --parameterfile FILENAME
File option: read input parameters from FILENAME
--c, --cmd Command line parameter option: read only boolean input parameters from command line and all other parameters will be assigned
from other sources
--l LOGFILENAME, --logfile LOGFILENAME
Output logfile name as specified
--d DIRECTORY_NAME, --directory DIRECTORY_NAME
Output directory name as specified
--version show program's version number and exit
--batch_mode, --bat Batch mode for plot. Otherwise interactive. (default: False)
Program flow¶
extract_desired_criteria: Extract desired refinement criteria
launch_segrefine3dcyclexplore_gui: Explore different refinement criteria interactively