Gallery¶
Spring3d¶
Segmentrefine3d¶
Program to iteratively refine 3D structure of helical specimens from segment stacks
Input: Image input stack refinement |
Output: Output volume name |
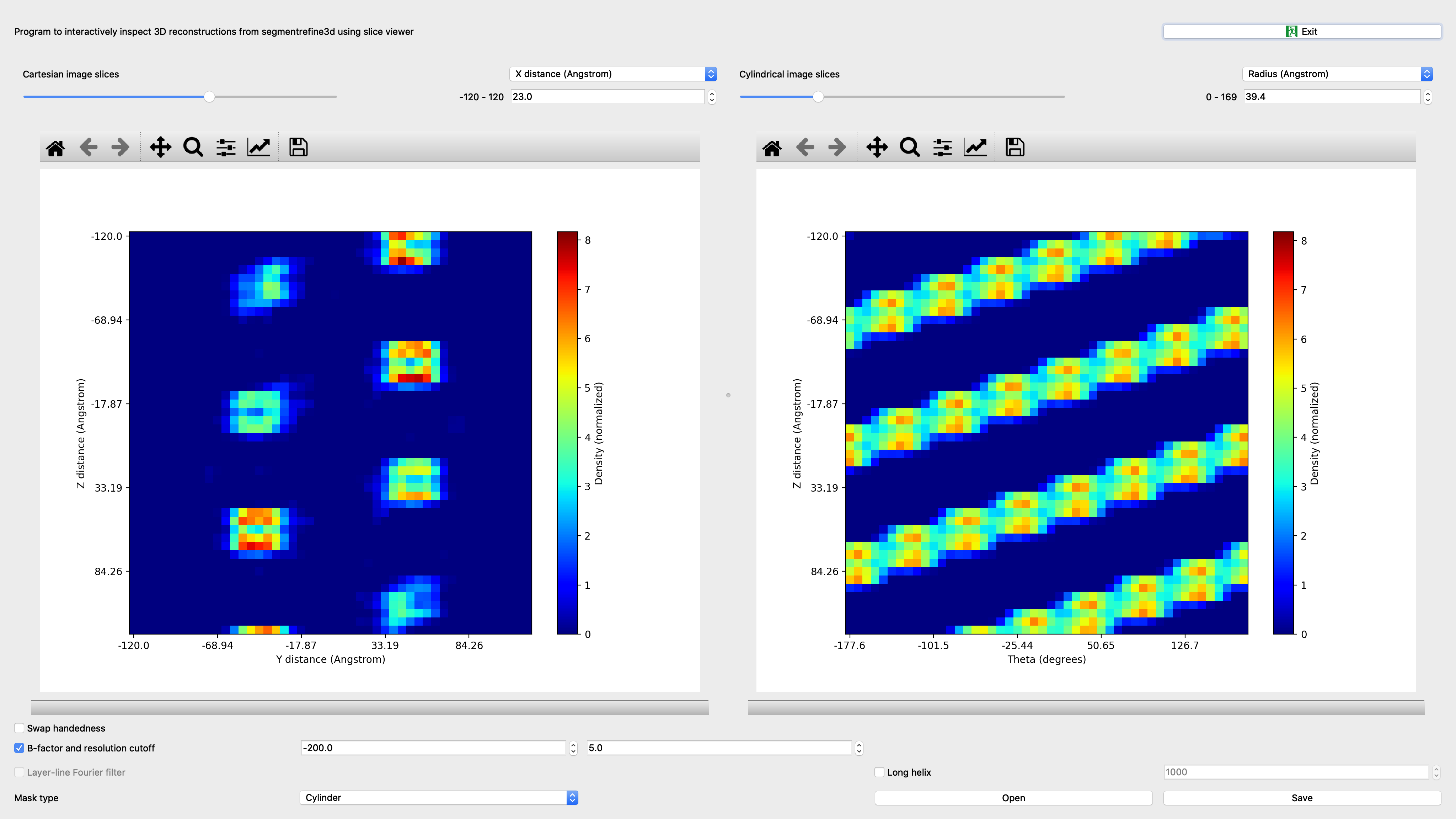
Segrefine3dinspect¶
Program to interactively inspect 3D reconstructions from segmentrefine3d using slice viewer
Input: Volume reconstruction |
Output: Interactive screenshot |
Segrefine3dgrid¶
Program to optimize segmentrefine3d reconstruction by varying refinement parameters systematically on a grid
Input: Image input stack refinement |
Output: Output volume name |
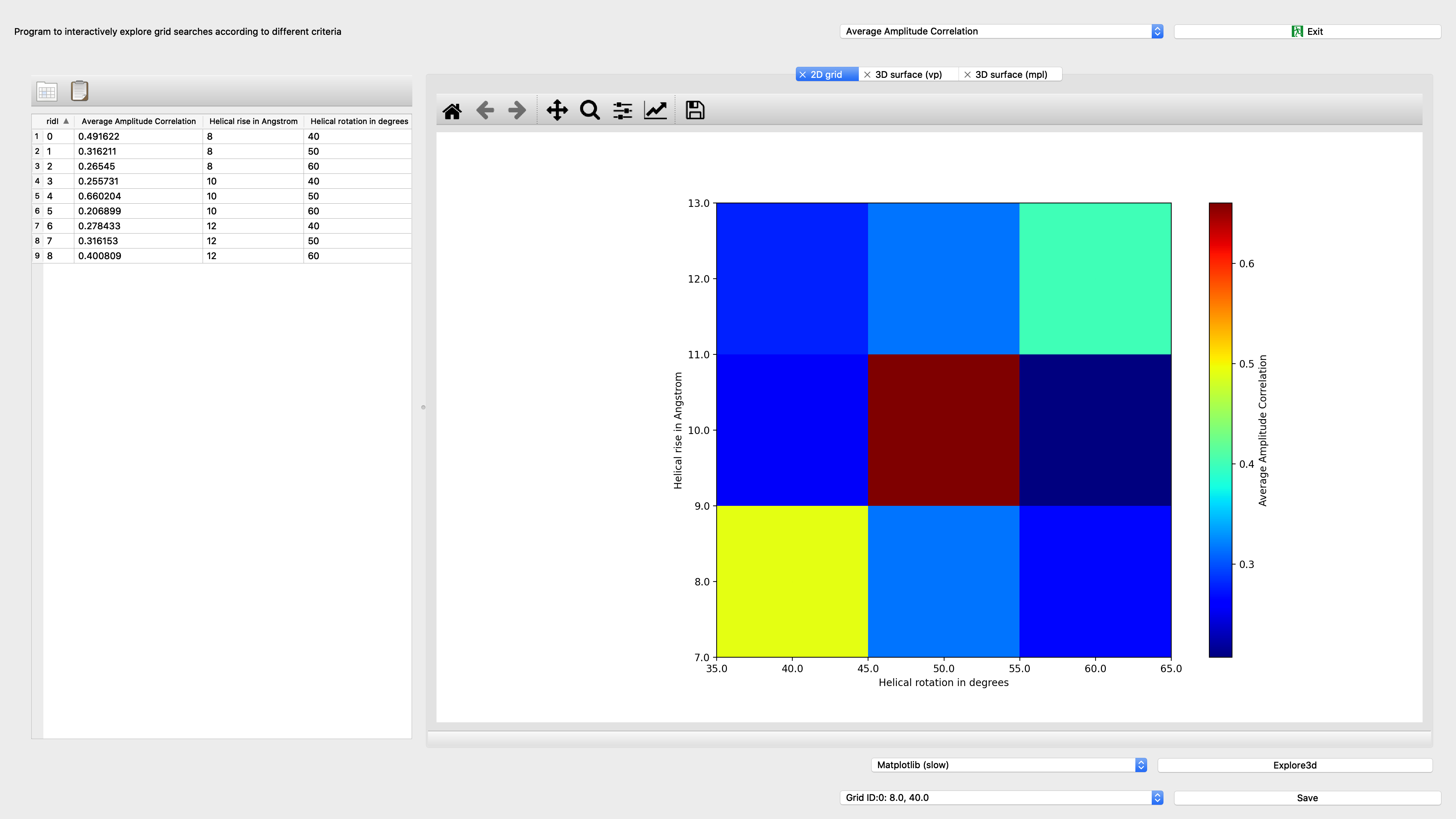
Seggridexplore¶
Program to interactively explore grid searches according to different criteria
Input: Grid database |
Output: Subgrid merge option |
Segmultirefine3d¶
Program to iteratively refine multiple 3D structures of helical specimens competitively from segment stacks
Input: Image input stack refinement |
Output: Output volume name |
Segclassreconstruct¶
Program to compute 3D reconstruction from a single class average using a range of different helical symmetries
Input: Class average stack |
Output: Volume name |
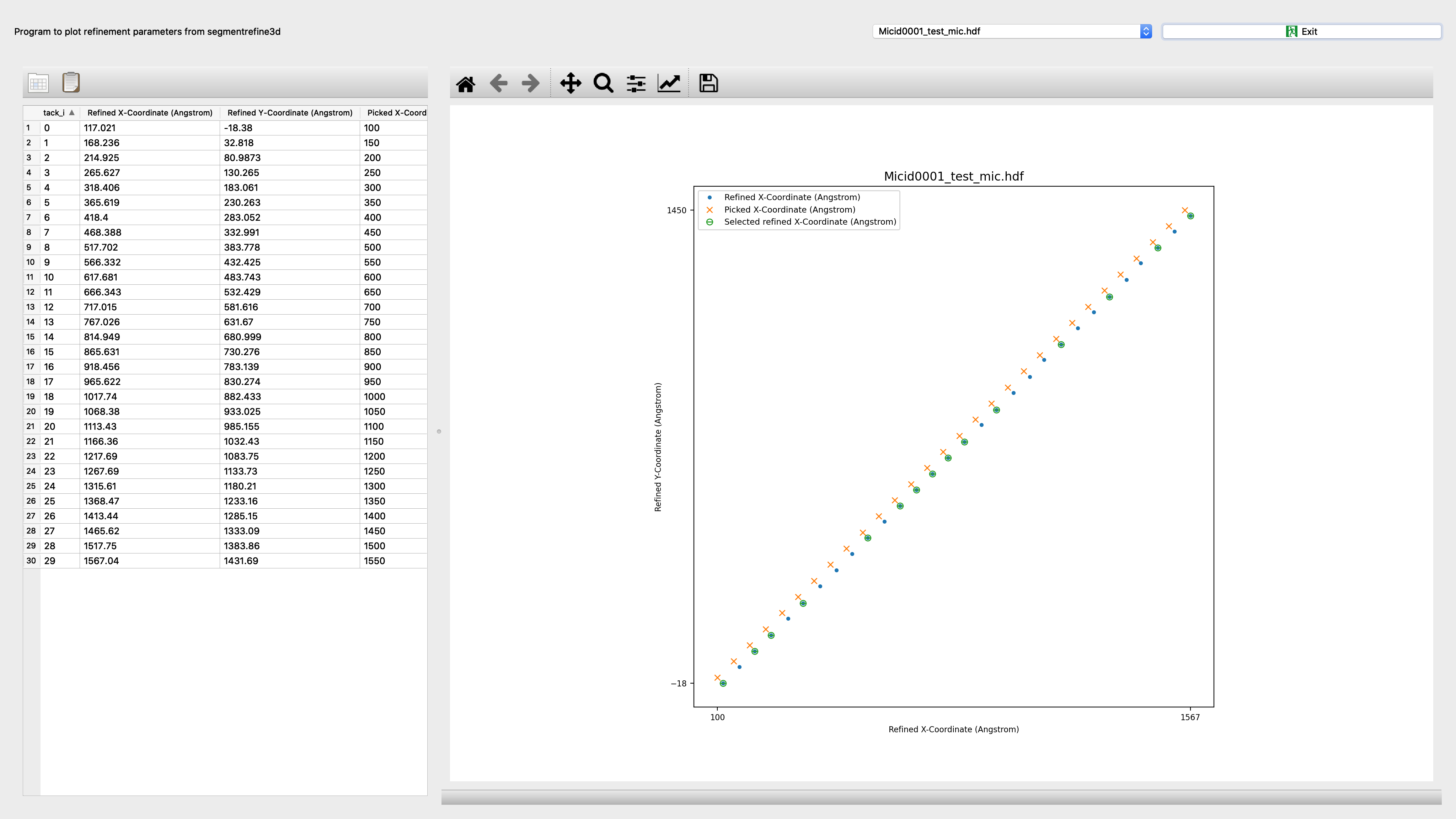
Segrefine3dplot¶
Program to plot refinement parameters from segmentrefine3d
Input: refinement.db file |
Output: Interactive screenshot |
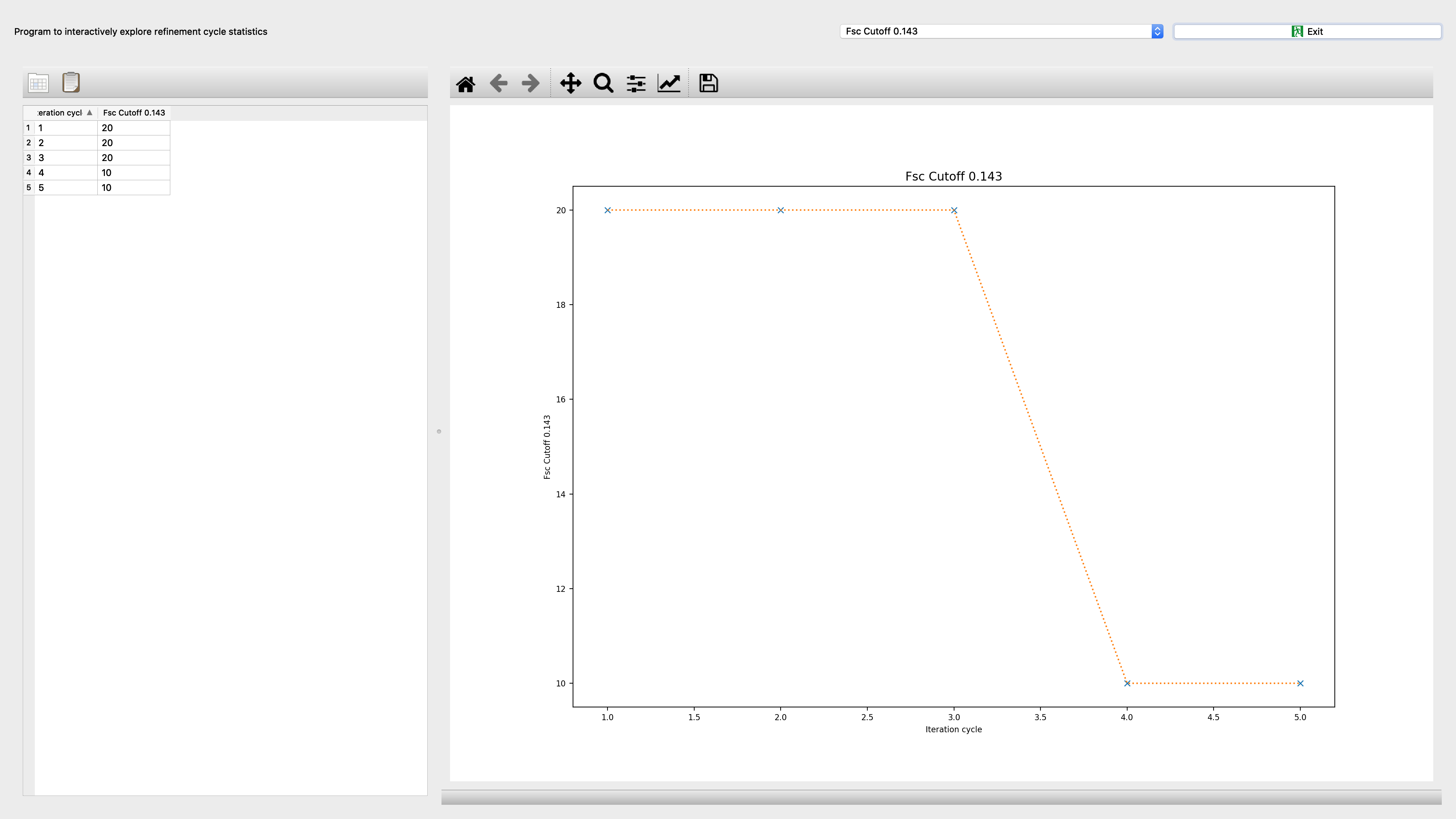
Segrefine3dcyclexplore¶
Program to interactively explore refinement cycle statistics
Input: refinement.db file |
Output: Interactive screenshot |
Segclassmodel¶
Program to assign a series of 3D models to class averages
Input: Reference volumes |
Output: Class average stack |
Spring2d¶
Segment¶
Program to extract overlapping segments from micrographs
Input: Micrographs |
Output: Image output stack |
Segmentexam¶
Program to examine all excised in-plane rotated segments and compute their collapsed (1D) and 2D power spectrum and width profile of helices
Input: Image input stack |
Output: Diagnostic plot |
Segmentclass¶
Program to classify excised in-plane rotated segments using SPARX’s k-means clustering
Input: Image input stack |
Output: Class average stack |
Segmentalign2d¶
Program to align segments from helical specimens with a restrained in-plane rotation of 0 or 180 +/- delta degrees
Input: Image input stack |
Output: Image output stack |
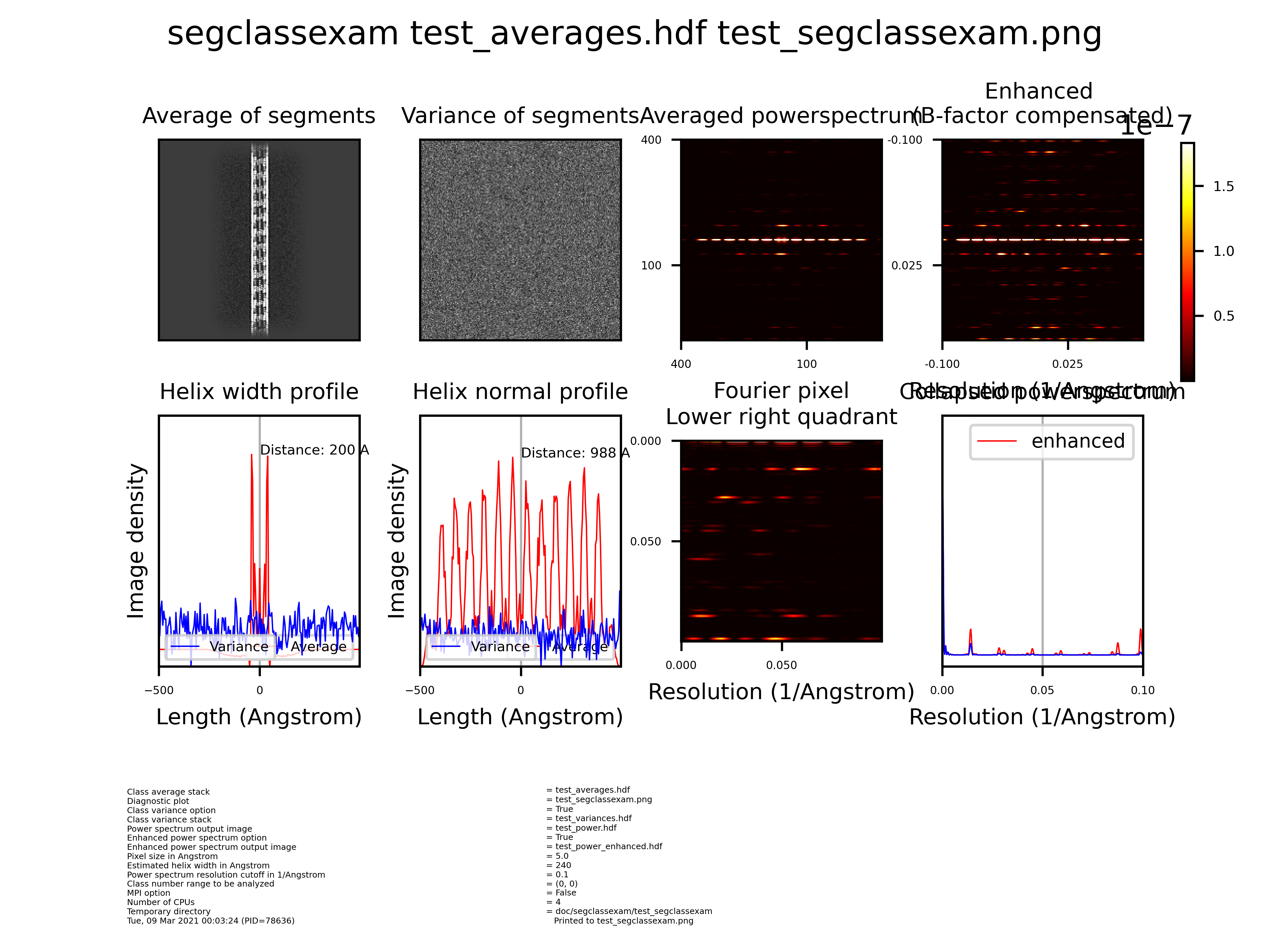
Segclassexam¶
Program to examine helix classes to compute their collapsed (1D) and 2D power spectrum and width profile of helices
Input: Class average stack |
Output: Diagnostic plot |
Segclasslayer¶
Program to extract amplitudes and phases from desired layer lines of class averages
Input: Class average stack |
Output: Interactive screenshot |
Seglayer2lattice¶
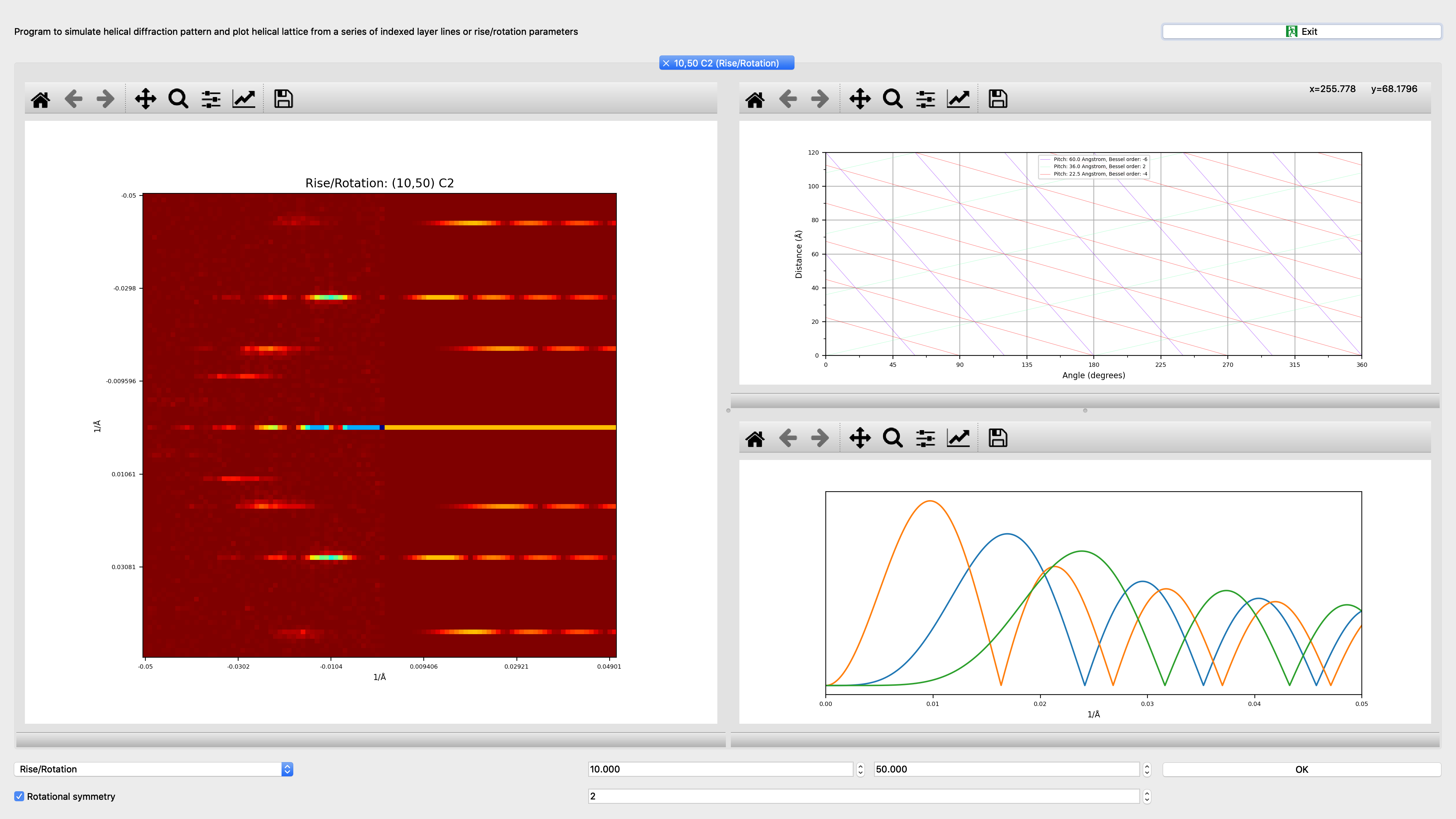
Program to simulate helical diffraction pattern and plot helical lattice from a series of indexed layer lines or rise/rotation parameters
Input: Analyze power spectrum |
Output: Power spectrum input image |
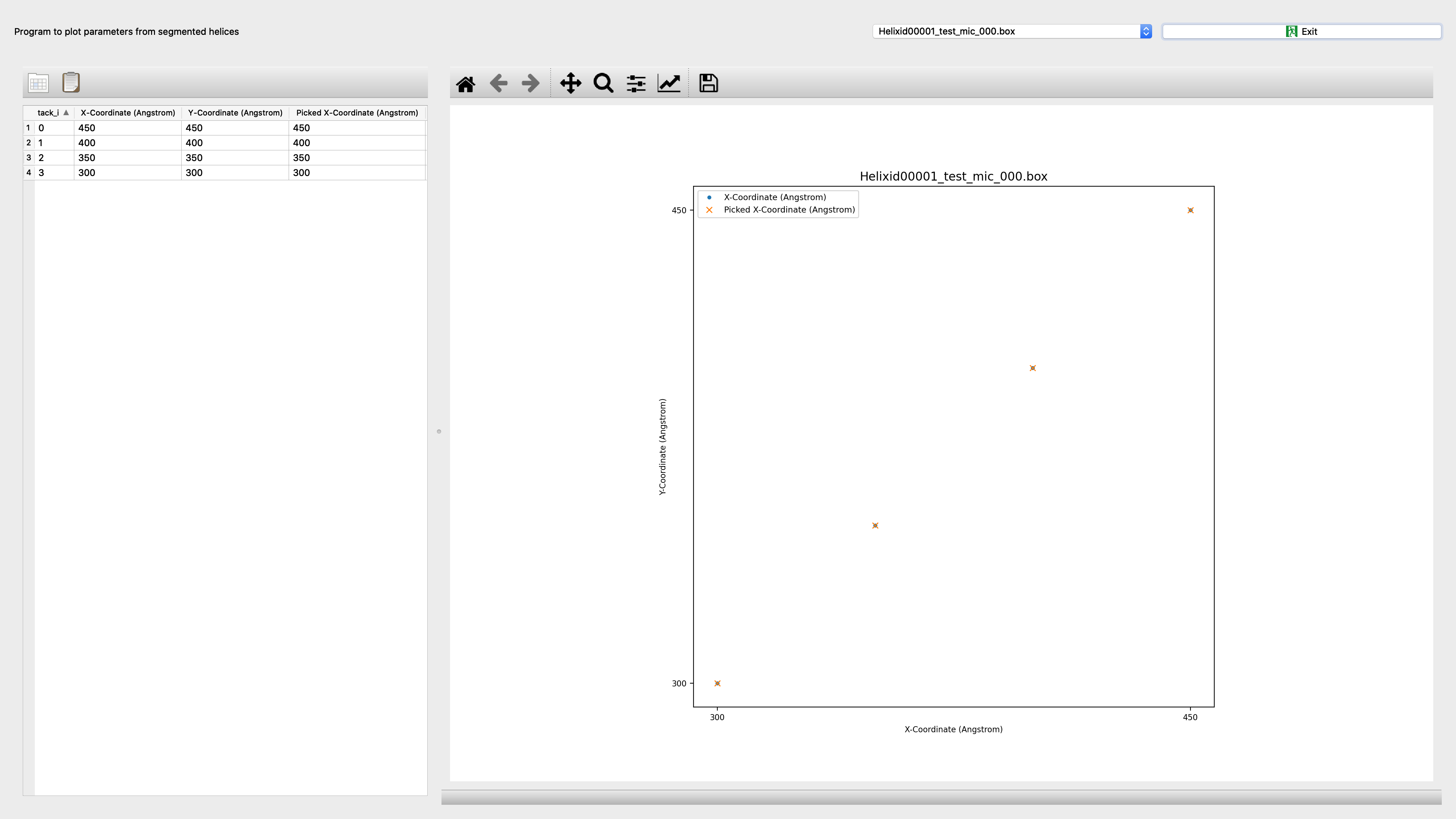
Segmentplot¶
Program to plot parameters from segmented helices
Input: spring.db file |
Output: Interactive screenshot |
Springmicrograph¶
Scansplit¶
Program to split large scan into individual micrographs. The location of individual micrographs is determined by a dummy micrograph reference.
Input: Micrographs |
Output: Output micrograph pattern |
Micexam¶
Program to examine micrograph quality by computing a localized power spectrum using EMAN2’s e2scaneval.py and an averaged power spectrum from overlapping tiles using SPARX’ sx_welch_pw2.py
Input: Micrographs |
Output: Diagnostic plot pattern |
Michelixtrace¶
Program to trace helices from micrographs
Input: Micrographs |
Output: Diagnostic plot pattern |
Michelixtracegrid¶
Program to trace helices from micrographs
Input: Micrographs |
Output: Diagnostic plot pattern |
Micctfdetermine¶
Program to determine CTF parameters from a set of micrographs using CTFFIND and CTFTILT (Mindell and Grigorieff JSB, 2003)
Input: Micrographs |
Output: Diagnostic plot pattern |
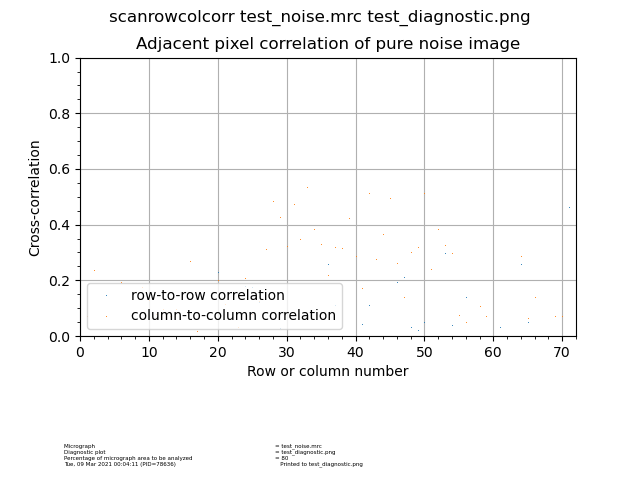
Scanrowcolcorr¶
Program to evaluate the performance of scanner by correlating adjacent rows and lines with each other from a pure noise image.
Input: Micrograph |
Output: Diagnostic plot |
Scandotfit¶
Program to evaluate performance of scanner by measuring dots spaced 2.5 mm apart.
Input: Micrograph |
Output: Diagnostic plot |
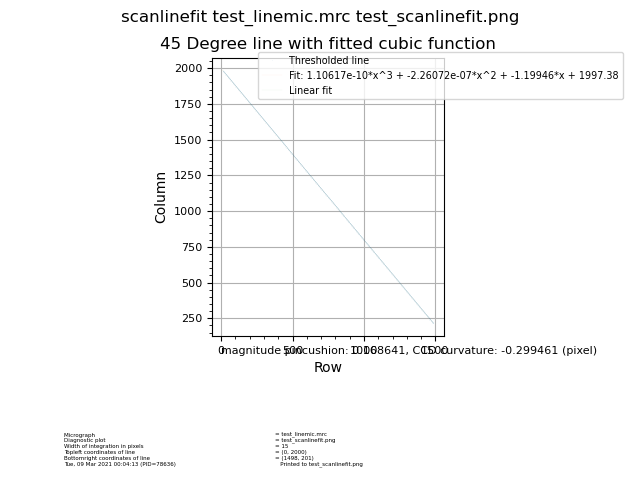
Scanlinefit¶
Program to evaluate scanner performance of scanner by measuring deviation from 45 degree line to determine CCD curvature and pincushion parameter.
Input: Micrograph |
Output: Diagnostic plot |